Compositions and methods for monitoring and altering protein folding and solubility
a technology of applied in the field of microorganisms, molecular biology and protein biochemistry, can solve the problems of inability to develop a robust assay for in vivo protein folding and solubility, and inability to detect the tendency of protein misfolding and aggrega
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials and Methods
[0251] Bacterial strains and plasmids. Wildtype E. coli strain MC4100 and a ΔtatC derivative of MC4100, strain B1LK0 (See, e.g., Bogsch et al., J Biol Chem 273, 18003-18006 (1998)), were used for all experiments. Plasmids for cytoplasmic expression of MBP (wt) and its folding mutant derivatives (See, e.g., Betton and Hofnung, J Biol Chem 271, 8046-8052 (1996)) were generated by inserting the gene encoding each MBP sequence into the NcoI / HindIII position of pTrc99A (Amersham Pharmacia). Plasmids for expressing MBP and its derivatives via the Tat pathway were created by excising the phoA gene from pTorA-AP (See, e.g., DeLisa et al., Proc Natl Acad Sci U S A 100, 6115-6120 (2003)) with XbaI and HindIII and inserting the gene encoding mature MBP (wt) or a folding mutant into the resulting XbaI / HindIII sites. Similarly, plasmids for localizing DsRed and its derivatives to the Tat pathway were generated by inserting either the DsRed gene sequence or its derivatives, ...
example 2
Folding Quality Control of the Tat Pathway
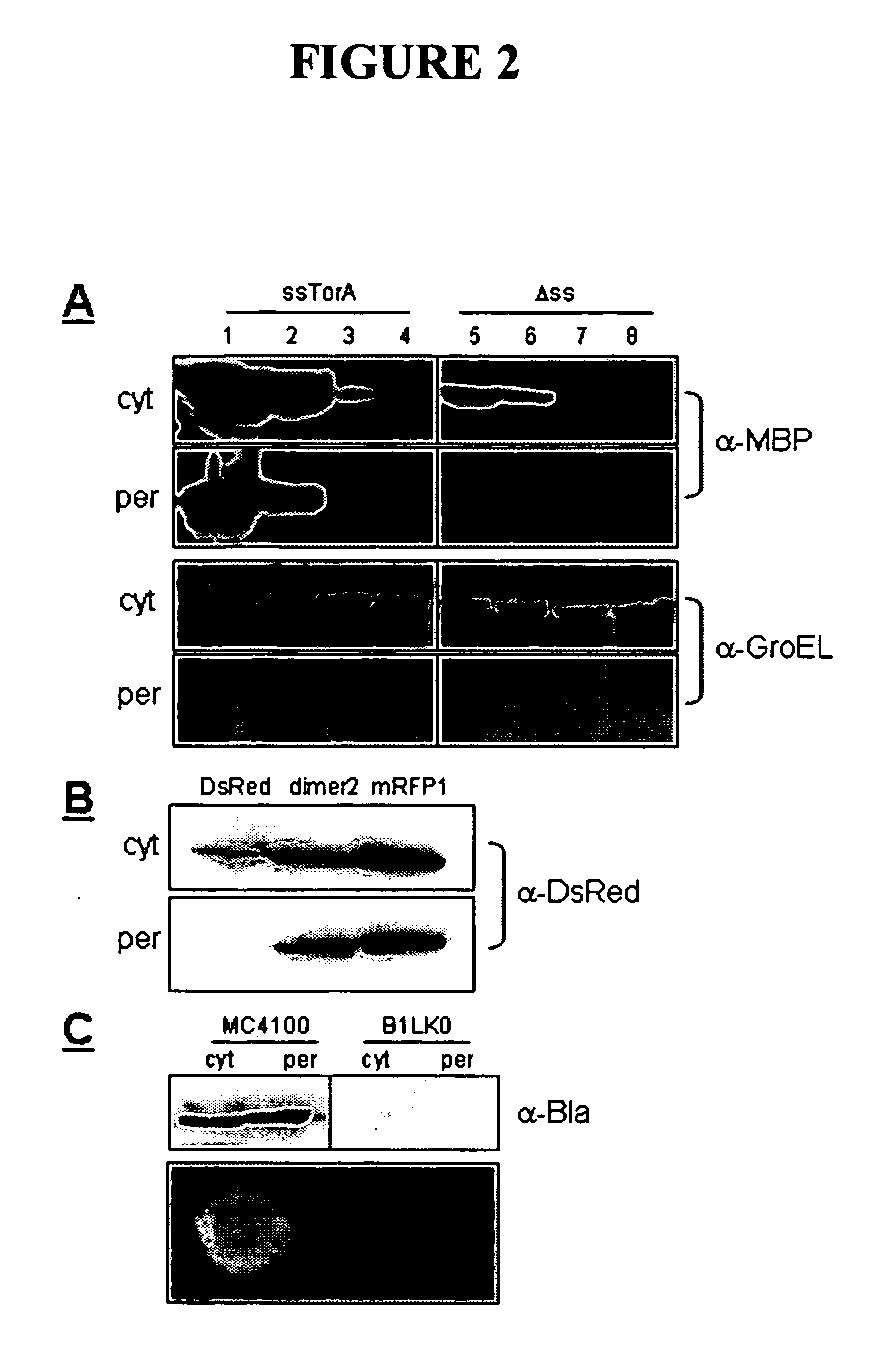
[0257] Tat transport of E. coli maltose binding protein (MBP) and three well-characterized MBP mutants prone to varying levels of off-pathway folding intermediates: MBP-G32D, MBP-I33P, and MalE31 (G32D / I33P) (See, e.g., Betton and Hoffnung, J Biol Chem 271, 8046-8052 (1996)) was evaluated. These proteins display a >100-fold difference in in vivo solubility with unfolding / refolding stability ranging from −5.5 kcal / mol to −9.5 kcal / mol ((See, e.g., Betton and Hofnung, J Biol Chem 271, 8046-8052 (1996)). The coding region for the well-characterized E. coli TMAO reductase twin-arginine signal peptide plus the first 4 residues of mature TorA (ssTorA, amino acids 1-46) (DeLisa et al., J Biol Chem 277, 29825-29831 (2002)) was fused upstream of the gene encoding the mature form of each MBP (residues 26-396), thus creating four ssTorA-MBP chimeras. Cell fractionation of wildtype MC4100 E. coli cells was performed to track subcellular localization an...
example 3
Tat-Based Solubility Reporter
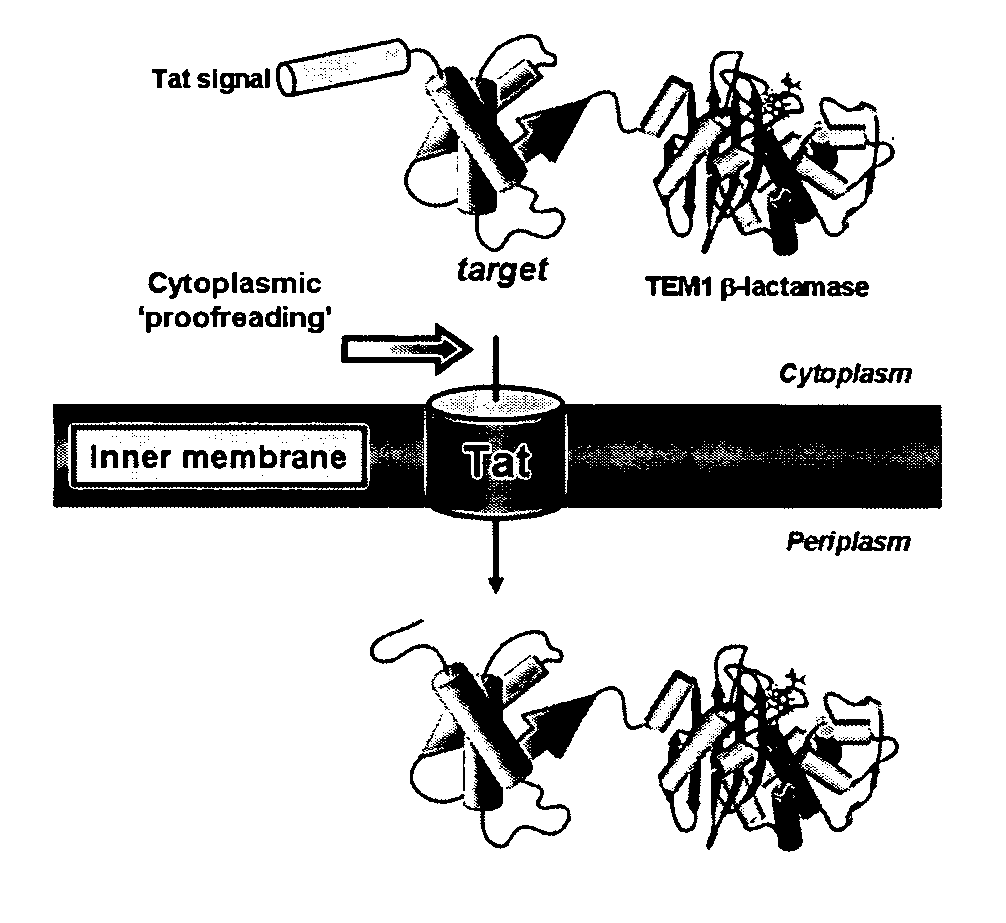
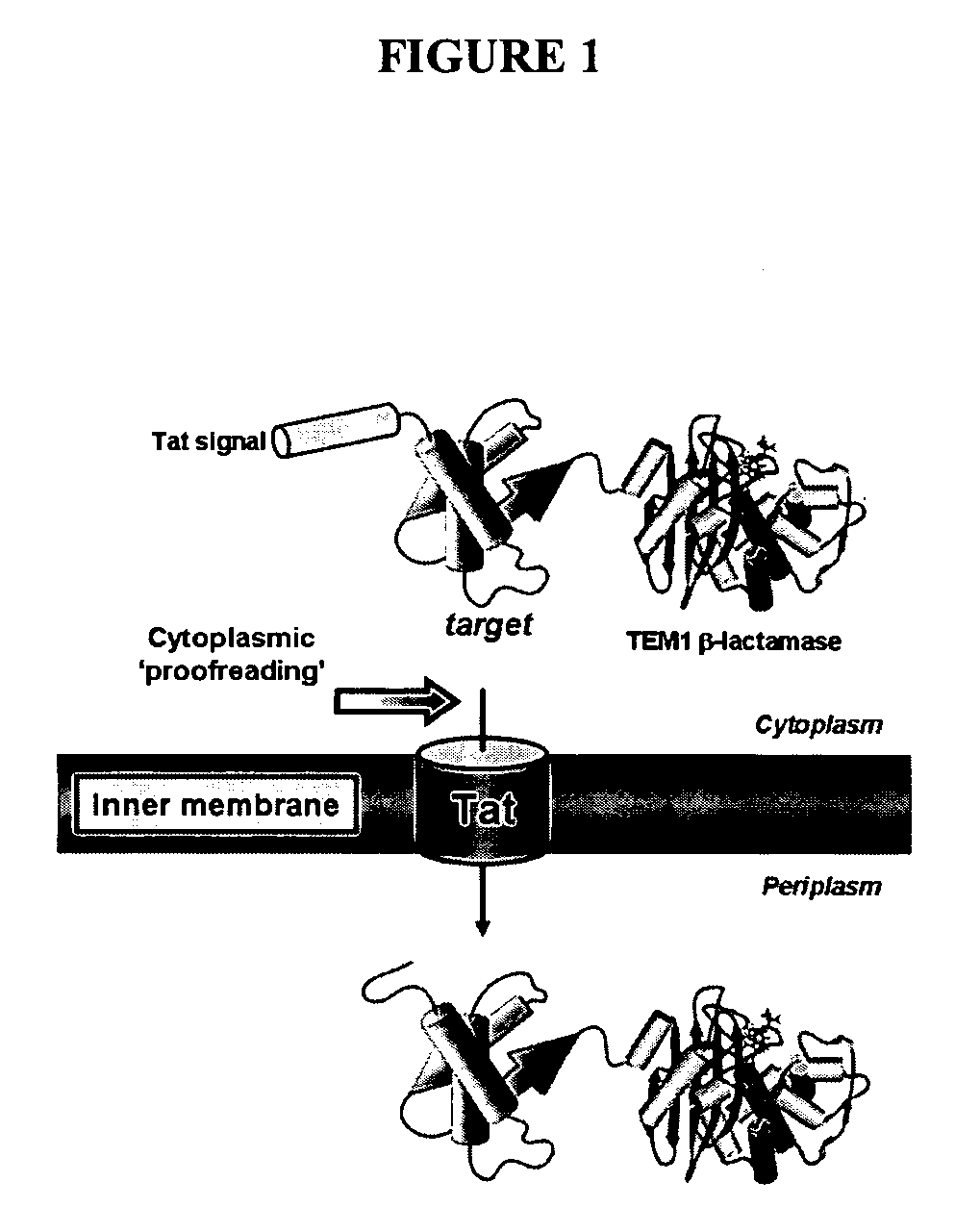
[0259] To exploit the quality control feature of the Tat pathway for monitoring protein solubility, a genetic assay that employs a tripartite fusion of the TorA signal peptide, a ‘target’ protein, and mature TEM1 β-lactamase (Bla) (FIG. 1A) was developed. The premise for this assay is as follows: a soluble target protein is exported to the periplasm via the Tat pathway and, by virtue of the Bla fusion, confers ampicillin resistance to E. coli cells expressing the ssTorA-target-Bla chimera. To verify that Bla is indeed capable of reporting Tat dependent transport in the assay, a vector (pTMB, FIG. 1B) was first constructed with no gene in the target position that expresses ssTorA-Bla. Upon expression of ssTorA-Bla in MC4100 and B1LK0, only periplasmic Bla localization was observed with a corresponding ampicillin resistance phenotype in MC4100 cells that possess a functional Tat pathway (FIG. 2C). Thus, Bla can be specifically transported by the Tat pathw...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com